Create Your First Project
Start adding your projects to your portfolio. Click on "Manage Projects" to get started
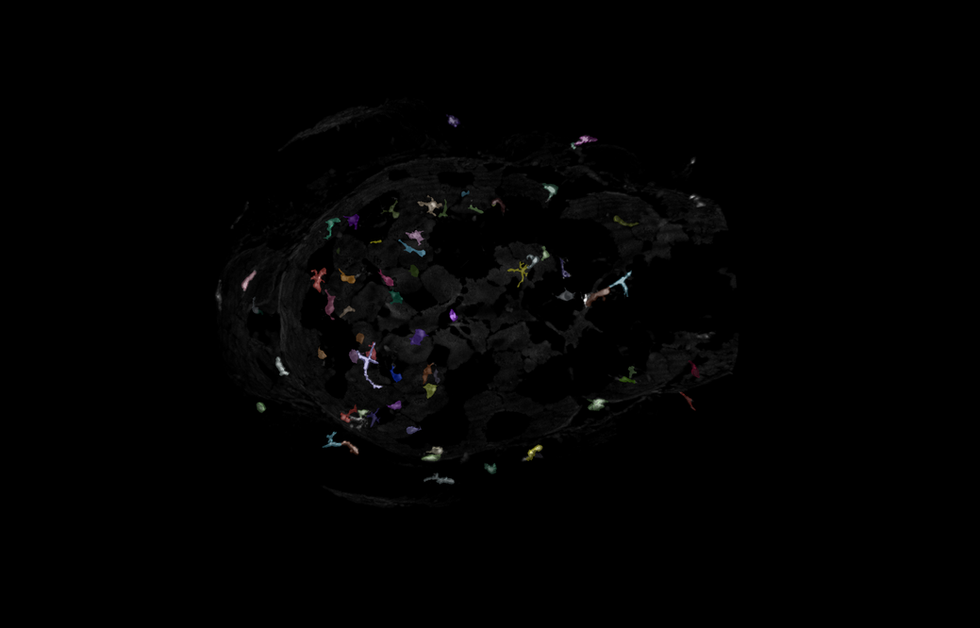
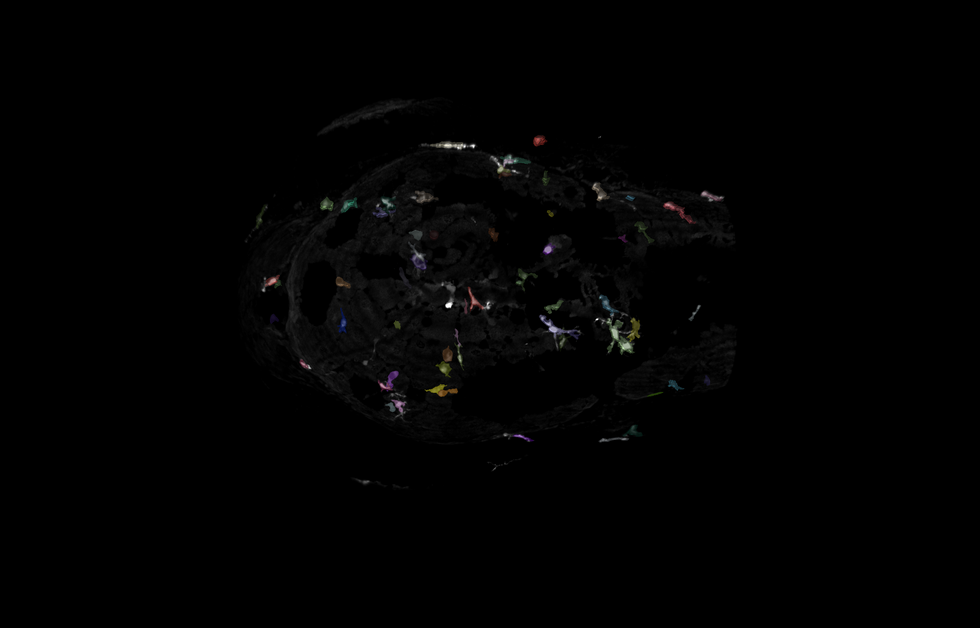
Zebrafish Microglia
The goal of this project is to accurately segment microglia in 3D from Z-stack images of zebrafish heads collected on a spinning disk confocal. Accurate segmentation of the microglia in 3D is critical to classifying the different states they present in. Some challenges with this data set included a very large z step, high background and inconsistent autofluorescence from other tissues in the samples.
I used transfer learning in CellPose's architecture to tune their cyto3 model to detect the fine protrusions and heterogeneous shapes of the microglia. Because of the large Z-step, training a 3D model in CellPose would be challenging for this dataset. To circumvent this, I leveraged training a 2D CellPose model with post-segmentation stitching to generate the final 3D objects.
A combination of imaris_ims_file_reader, pyclesperanto (GPU accelerated image processing), CellPose, numpy, scikit-image, and pandas was used to created a reproducible pipeline to analyze this data set in batch.